Genome-wide quantitative enhancer activity maps identified by STARR-seq
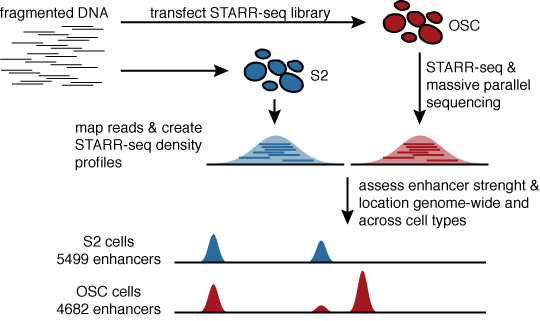 Genomic enhancers are important regulators of gene expression, but their identification is a challenge and methods depend on indirect measures of activity. We developed a method termed STARR-seq to directly and quantitatively assess enhancer activity for millions of candidates from arbitrary sources of DNA, enabling screens across entire genomes. When applied to the Drosophila genome, STARR-seq identifies thousands of cell type-specific enhancers across a broad continuum of strengths, linking differential gene expression to differences in enhancer activity and creating a genome-wide quantitative enhancer map. This map reveals the highly complex regulation of transcription, with several independent enhancers for both developmental regulators and ubiquitously expressed genes. STARR-seq can be used to identify and quantitate enhancer activity in other eukaryotes, including human.
Genomic enhancers are important regulators of gene expression, but their identification is a challenge and methods depend on indirect measures of activity. We developed a method termed STARR-seq to directly and quantitatively assess enhancer activity for millions of candidates from arbitrary sources of DNA, enabling screens across entire genomes. When applied to the Drosophila genome, STARR-seq identifies thousands of cell type-specific enhancers across a broad continuum of strengths, linking differential gene expression to differences in enhancer activity and creating a genome-wide quantitative enhancer map. This map reveals the highly complex regulation of transcription, with several independent enhancers for both developmental regulators and ubiquitously expressed genes. STARR-seq can be used to identify and quantitate enhancer activity in other eukaryotes, including human.
Arnold CD, Gerlach D, Stelzer C, Boryń LM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 2013 Mar 1;339(6123):1074-1077. Pubmed 23328393.
Reviews and Coverage
- Louisa Flintoft. Gene regulation: Enhancing the hunt for enhancers. Nature Reviews Genetics 14, 151 (2013).
- Beth Moorefield. STARR-seq enterprise. Nature Structural & Molecular Biology 20, 308 (2013).
- No Author. Genome-wide enhancer maps. Nature Methods 10, 193 (2013).