Our genome-scale characterization of in vivo activities for 7705 enhancer candidates throughout Drosophila embryogenesis is published in Nature [1].
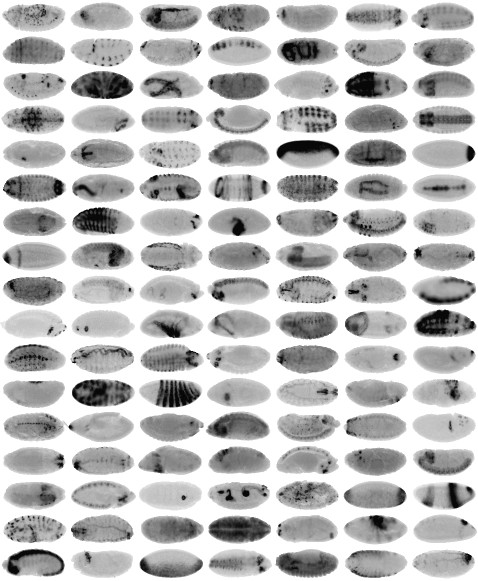 Transcriptional enhancers are crucial regulators of gene expression and animal development and the characterization of their genomic organization, spatiotemporal activities and sequence properties is a key goal in modern biology. Here we characterize the in vivo activity of 7,705 Drosophila melanogaster enhancer candidates covering 13.5% of the non-coding non-repetitive genome throughout embryogenesis. 3,557 (46%) candidates are active, suggesting a high density with 50,000 to 100,000 developmental enhancers genome-wide. The vast majority of enhancers display specific spatial patterns that are highly dynamic during development. Most appear to regulate their neighbouring genes, suggesting that the cis-regulatory genome is organized locally into domains, which are supported by chromosomal domains, insulator binding and genome evolution. However, 12 to 21 per cent of enhancers appear to skip non-expressed neighbours and regulate a more distal gene. Finally, we computationally identify cis-regulatory motifs that are predictive and required for enhancer activity, as we validate experimentally. This work provides global insights into the organization of an animal regulatory genome and the make-up of enhancer sequences and confirms and generalizes principles from previous studies. All enhancer patterns are annotated manually with a controlled vocabulary and all results are available through a web interface http://enhancers.starklab.org, including the raw images of all microscopy slides for manual inspection at arbitrary zoom levels.
Transcriptional enhancers are crucial regulators of gene expression and animal development and the characterization of their genomic organization, spatiotemporal activities and sequence properties is a key goal in modern biology. Here we characterize the in vivo activity of 7,705 Drosophila melanogaster enhancer candidates covering 13.5% of the non-coding non-repetitive genome throughout embryogenesis. 3,557 (46%) candidates are active, suggesting a high density with 50,000 to 100,000 developmental enhancers genome-wide. The vast majority of enhancers display specific spatial patterns that are highly dynamic during development. Most appear to regulate their neighbouring genes, suggesting that the cis-regulatory genome is organized locally into domains, which are supported by chromosomal domains, insulator binding and genome evolution. However, 12 to 21 per cent of enhancers appear to skip non-expressed neighbours and regulate a more distal gene. Finally, we computationally identify cis-regulatory motifs that are predictive and required for enhancer activity, as we validate experimentally. This work provides global insights into the organization of an animal regulatory genome and the make-up of enhancer sequences and confirms and generalizes principles from previous studies. All enhancer patterns are annotated manually with a controlled vocabulary and all results are available through a web interface http://enhancers.starklab.org, including the raw images of all microscopy slides for manual inspection at arbitrary zoom levels.
[1] Kvon EZ, Kazmar T, Stampfel G*, Yáñez-Cuna JO*, Pagani M, Schernhuber K, Dickson BJ, Stark A. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature, 2014. Epub ahead of print. Pubmed 24896182.